1.0
1.0
Webserver for Simulation of Diffusional Association (SDA): making life much easier to
simulate macromolecular diffusion.
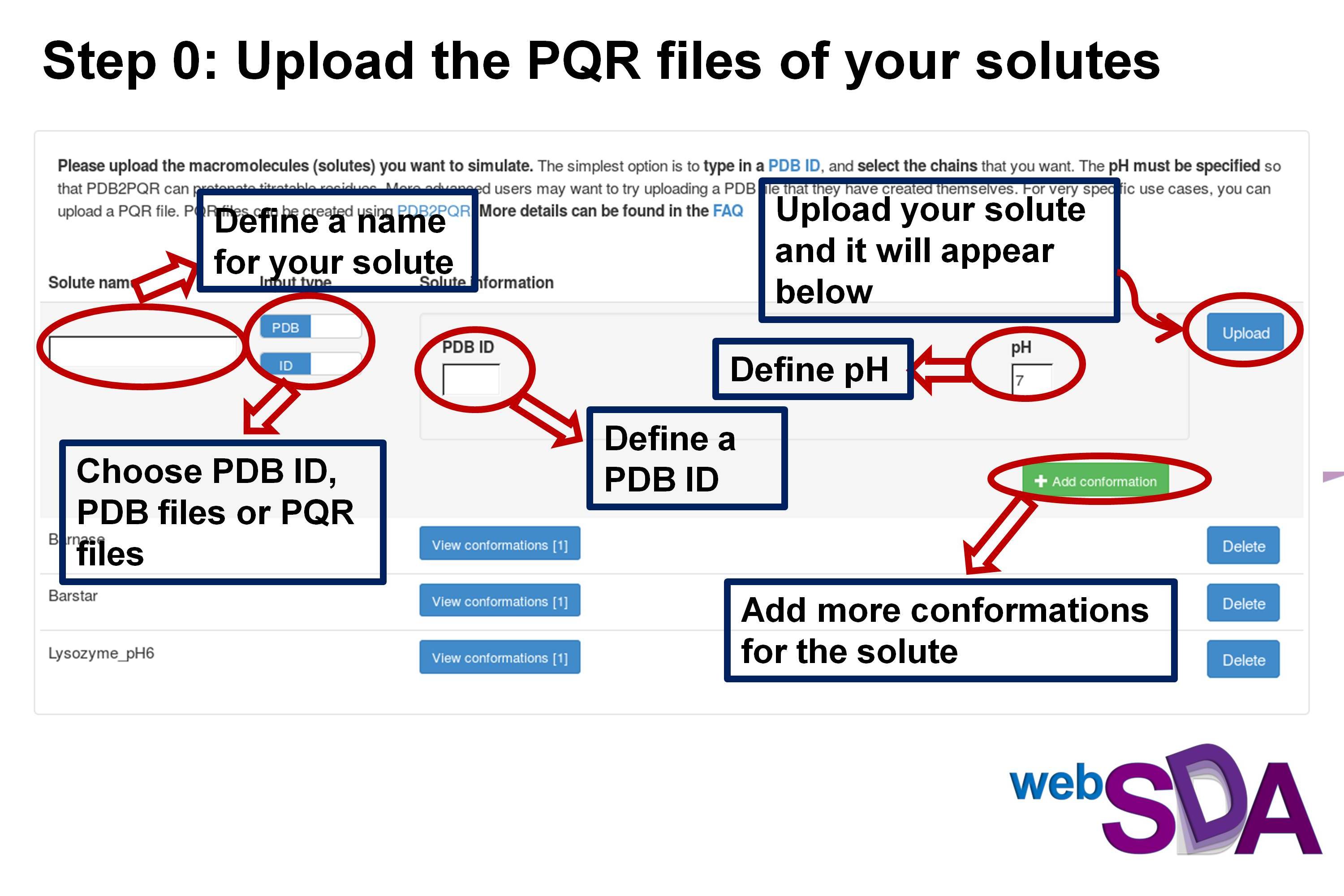
Step 0: Upload your solutes for the simulation
Please upload the macromolecules (solutes) you want to simulate. The simplest option is to type in a PDB ID, and select the chains that you want. The pH must be specified so that PDB2PQR can protonate titratable residues. More advanced users may want to try uploading a PDB file that they have created themselves. For very specific use cases, you can upload a PQR file. PQR files can be created using PDB2PQR. More details can be found in the FAQ
You do not have any solutes uploaded.
You need to have at least two solute for SDA docking and association and at least one solute for sdamm.
Or you can try the examples files: Barnase.pqr and Barstar.pqr for "SDA docking" and "SDA association", and Lysozyme_pH6.pqr for "SDA multiple molecules"
Load example files